TL;DR
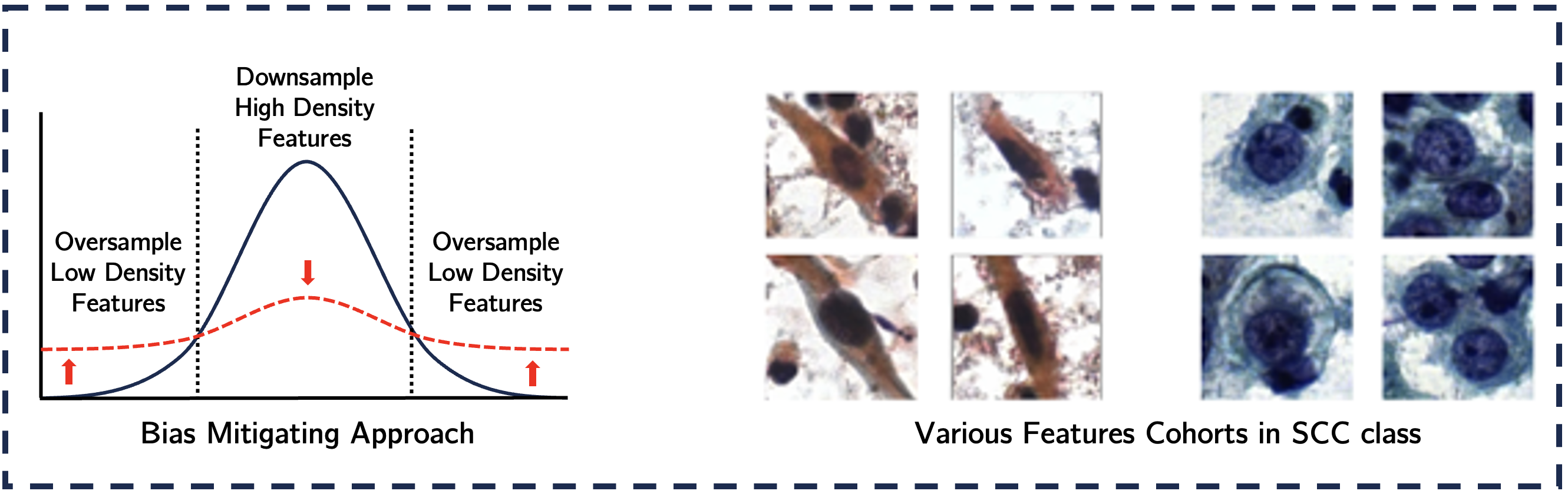
We introduce a bias-mitigating approach that categorizes data points into high- and low-density feature cohorts based on feature probability distributions. Using this sampling based approach, we provide a balanced signal to the classifier for an 8-way classification task. We demonstrate that oversampling low-density cohorts during training significantly reduces AUC disparity between cohorts by 73.42%. This results in improved diagnostic accuracy and consistency in DL-based cervical cytology workflows.
Abstract
Cervical cancer poses a serious health risk to women all across the world. The advancements in deep learning (DL), have driven the rise of DL-assisted cervical cytology screening methods for various diagnostic tasks. However, most of these DL approaches are susceptible to the inherent bias in clinical datasets, which restricts their practical deployment. A key source of bias in DL-based cervical cytology workflows is the high variability in the representation of various features extracted by these models across different classes. This imbalanced feature representation results in inconsistent model performance across different feature cohorts, which is known as feature bias. Building on this understanding, our work underscores the importance of mitigating feature bias in DL-based cervical cytology workflows. We demonstrate that effective bias mitigation reduces skewness in performance metrics, which improves diagnostic performance and enhances patient outcomes.
Methodology
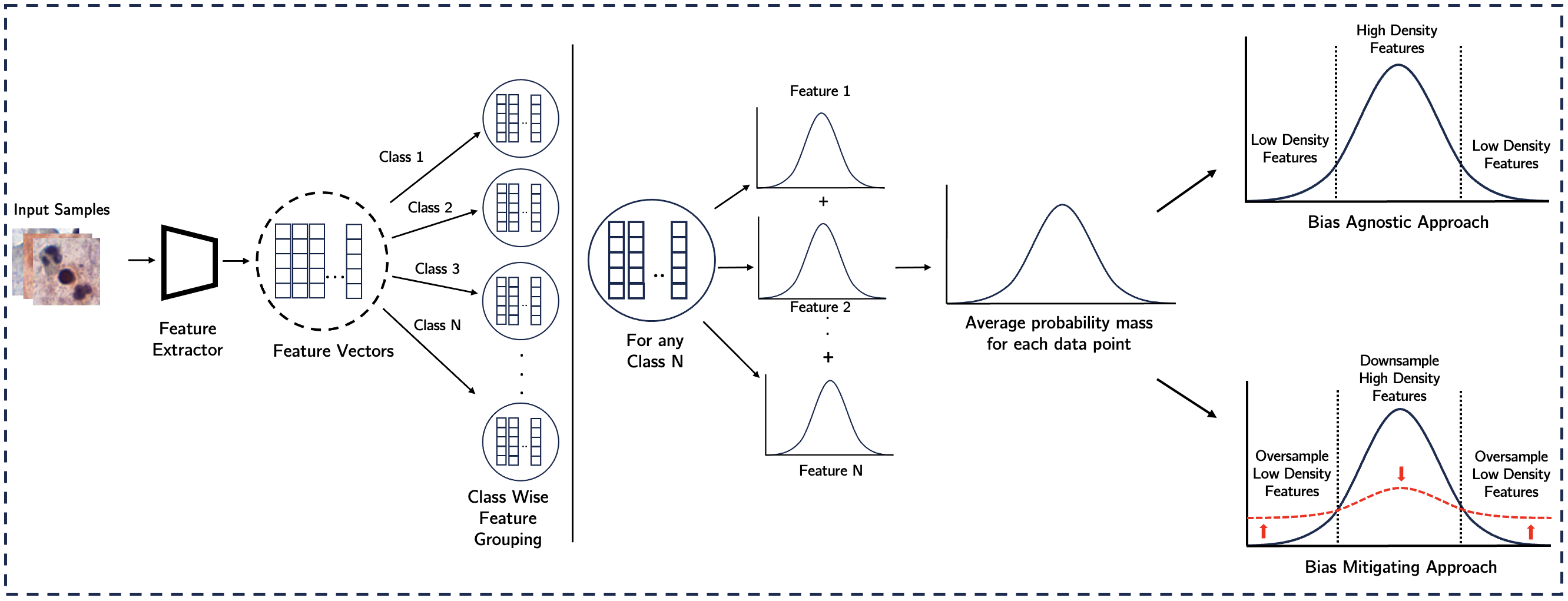
Our approach uses DieT as a feature extractor to generate D-dimensional feature vectors, which are segregated class-wise. For each class, we compute the probability distribution across feature dimensions, calculate the average probability mass for each data point, and categorize data into high-density and low-density cohorts. Then, the model is trained using bias-agnostic and bias-mitigating approaches, with performance evaluated on both cohorts using AUC as the performance metric. While the bias-agnostic approach relies on random sampling, leading to a dominance of high-density features during training. Conversely, the bias-aware approach ensures parity by oversampling low-density feature cohorts (vice-versa for high-density feature cohorts), allowing the model to receive a balanced signal from both cohorts.
Results
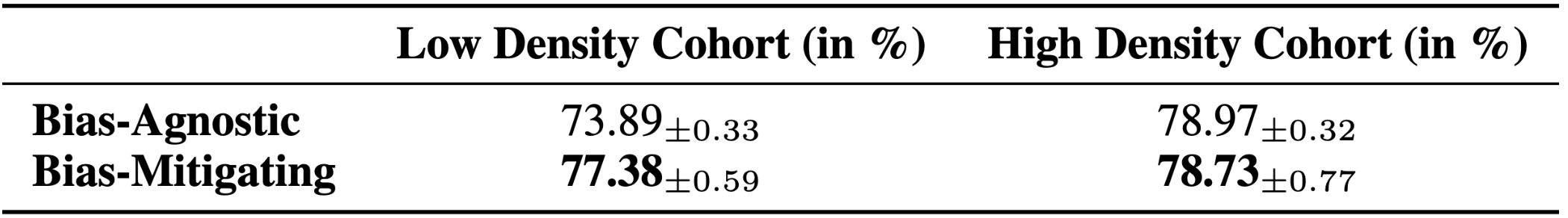
Our results shows that the Bias-Agnostic approach yields an AUC difference of 5.08% between the two cohorts of the CRIC dataset compared to the Bias-Mitigating model which obtains an AUC difference of 1.35%. This represents a 73.42% reduction in the AUC difference compared to the Bias-Agnostic approach. Our results demonstrate that using an effective bias-mitigating approach reduces the skewness in the model performance across various feature cohorts. These results can improve diagnostic accuracy and consistency in DL-based cervical cytology workflows.